Projects
Current/Ongoing Projects
(1) A network-based blueprint for cellular organization
Membraneless, subcellular compartments (‘condensates’) form by biological phase transitions. Condensates have been implicated in RNA processing and signaling, but function for most remains unclear. We use quantitative cellular reconstitution approaches, which rely on custom methods, to uncover the rules that dictate their unique compositions and functions. Our published research used two closely related RNA-rich cytoplasmic condensates—stress granules and processing bodies—as prototypes to inform a blueprint for intracellular patterning in the absence of membranes. We found that network connectivity between essential oligomeric ‘nodes’ (determines the molecular components of condensates and whether they adhere, mix, or disassociate. Remarkably, proteins that lack network connectivity ‘cap’ such nodes and break the required connectivity for stress-dependent condensation. We predict that such ‘valence capping’ is a general principle that controls directional substrate processing, signaling pathway rewiring, and gradient-based patterning in development (see Project 3). Importantly, peptides and small molecule caps represent a new network-based approach for drug design. Current and future work focuses on the function of molecular networks in development (e.g., myocyte differentiation) and how such networks are rewired by protein nodes and caps in diseases of aging (neurodegeneration, myopathies, cancers).
(2) Vertebrate-specific compartments in RNA homeostasis
Most membraneless compartments have unclear function. Acutely prominent amongst these are ‘speckles’, which cover 15% of the human cell nucleus and are present in all vertebrates but absent in simple eukaryotes (yeast, worms). Organisms with speckles have complex mRNA processing programs, which may inform function. An extreme example is that of skeletal muscle cells , which rewire mRNA processing during differentiation to stitch (‘splice’) the huge mRNAs encoding their structural proteins. To determine speckle function, methods that turn speckles on (‘assembly’) and off (‘disassembly’) are needed.
In work that is soon to be pre-printed, we used a suite of orthogonal experimental approaches to address this need. With an unbiased CRISPR screen, we discovered a single node uniquely essential for the formation of speckles. Serindipitously, David’s unpublished work from grad school on the interactome of tau aggregates implicated the same node. This gigantic protein, SRRM2, is disordered but for a conserved oligomerization domain that fits into the spliceosome’s catalytic groove. As splicing complexity increased and speckles evolved, its RNA-binding serine-arginine (SR) repeats expanded. Reconstitution approaches have revealed a necessary role for multimerized intrinsically disordered regions (IDRs) in speckle assembly. Based on unexpected similarities between tau aggregate-binding proteins, we discovered peptide caps that dissolve speckles and aggregates. We engineered this cap so that physiological speckles are spared while toxic tau assemblies are eliminated. This work represents powerful proof-of-principle for therapeutic caps designed to break pathological networks (see Project 3), and inform a new molecular model for common neurodegenerative diseases.
We are now working to provide an evolutionary basis for nuclear speckles, in addition to clinically minded endeavors. We aim to determine their function in regulating RNA homeostasis, tackling a series of hypothesis-driven questions made possible by our new methods. In, we are using published approaches to control stress granule assembly to test whether they act as a secondary defense against failures in RNA homeostasis.
(3) Network deconstruction for cap discovery in health and disease
Proteins with oligomerization domains (‘ODs’) are frequently implicated in signaling cascades and diseases of aging (e.g., neuromuscular, cancers). Our network model predicts that ODs are over-represented in disease and signaling, by virtue of their unique switch-like control over network wiring and biological reactions. However, the binding interfaces on ODs that are critical to this switch are rarely targets of drug discovery, nor are binding partner stoichiometries appreciated in signaling. We envision that knowledge of cap-node binding interfaces and the molecules that target them will inform both drug design and signaling pathways in development. Our unpublished work closes this gap, with the goal of targeting ODs to understand health and develop new therapies for disease. We used dozens of ODs to demonstrate proof-of-concept for a discovery pipeline that can identify the proteins (caps, nodes) that exert direct control over molecular networks. We plan to use this pipeline to dismantle physiological and pathological networks into ‘parts’ lists for mechanistic interrogation of developmental pathways (skeletal muscle formation) and diseases of aging.
(4) Cell-cell fusion: Finding patterns in multinucleated cells
Multinucleated cells form by cell-cell fusion or division without cytokinesis. Such ‘syncytia’ are critical to development, comprise much of the human body, and are hijacked by viruses to escape the immune response. A prominent example is skeletal muscle, whose fibers feature cell type-specific condensates, displays exquisite patterning, and degenerate in disease (‘myopathies’). The mechanistic basis for untreatable myopathies is unknown but could result from failures in patterning and rejuvenation. We envision that node and cap gradients are central to all-or-none signaling pathways in differentiated cells, forming Turing patterns.
To test this, David developed a highly precise method to fuse cells on demand, generating molecular gradients at specific locations. We are applying this method to a model of skeletal muscle differentiation (IPSCs) to dissect the role of patterned condensation in development and disease.
(5) Unleashing the methodological power of Optogenetics
New methods and unexpected collaborations are what drives great scientific discoveries. To this end, we have developed optogenetic tools to bring order to chaotic molecular networks. These tools not only allow rigorous testing of phase transition hypotheses in living cells, but also have predictive power for cellular biology beyond condensates., with broad applicability for diverse questions (from signaling to enzymatic reactions; development to disease; cancer to neurodegeneration). Specifically, our condensate-based assays can be applied to any system in which a crosslink (e.g., protein-protein interaction) is available, allowing unprecedented spatiotemporal control over molecular interactions and signaling pathways in living systems.
We have developed synthetic optogenetic platforms to
A. Reconstitute physiological, multi-component condensates from minimal domain modules
B. Assess relative binding affinity of protein-protein interactions in living cells (Opto-PPI)
C. Illuminate the role of ligand competition (‘nodes’ vs. ‘caps’) in cellular phase transitions
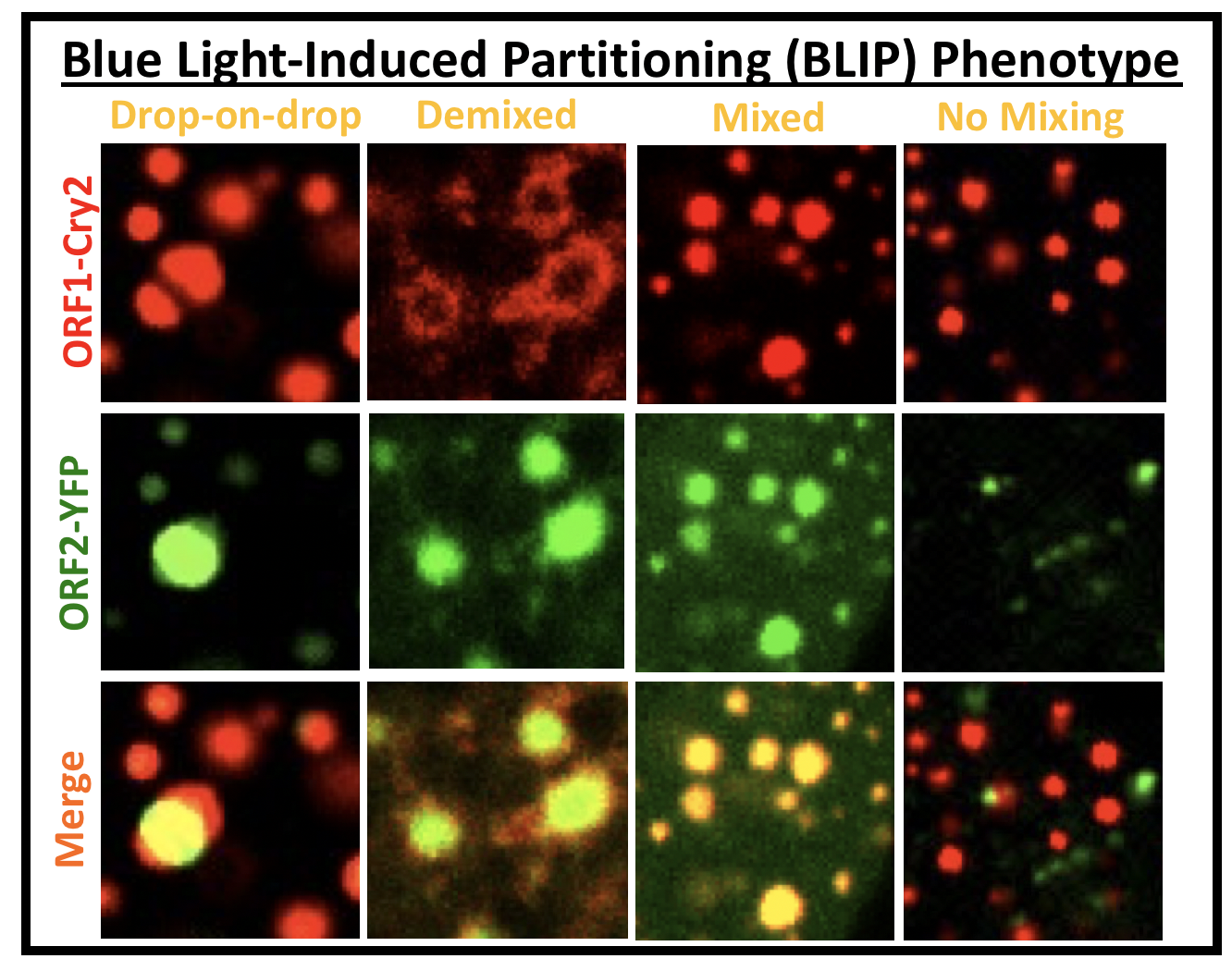
D. ‘Cheat’ the diffraction barrier to probe the composition of multiphase condensates (BLIP; see image panel)
Currently, we are optimizing/developing new methods to
E. Increase throughput and reliability of proximity-based proteomics (Opto-ID)
F. Control cell-cell fusion (Opto-FUS…just to confuse fans of fused in sarcoma and focused ultrasound)
G. Study whatever we get excited about tomorrow (Opto-Grants)
The past is not even past: Rejuvenated projects
(6) Tau strains: Self-replicating structures encode diseases
In David’s PhD research at Washington University (advisor: Marc Diamond), he pursued the molecular origin of tauopathies (Alzheimer’s, frontotemporal degeneration, etc.), which are neurodegenerative diseases caused by aggregates of the tau protein. Despite this similarity, each disease features a unique clinical and pathological profile. He discovered that individual tauopathies are true disease strains, specified by isolatable and self-replicating tau aggregate structures. These findings inform ongoing development of structure-based therapeutics to halt the spread of neurodegeneration and highlight a remarkable ability for protein assemblies to encode information. Currently, the lab is circling back to this foundational work, creating new cellular models to understand the origin and toxicity (or….lack-there-of) of protein aggregates (e.g., TDP-43, ASyn, TMEM106B, ANXA11) in neurodegenerative diseases. We remain fixated on potential functional, self-propagating, higher-order assemblies (or ‘prions’).
SEE PUBLICATIONS FOR MORE INFORMATION!